Week 7 - Bioinformatics
Bioinformatics is the application of computer technology to the management of biological information. Computers:
- store
- gather
- analyze
- integrate
all genetic information which can be applied to gene-based drug discovery and development.
Transcription
DNA sequences are composed of the 4 bases:
- Adenine (A)
- Thymine (T)
- Guanine (G)
- Cytosine (C)
A's are matched with T's and C's w/ G's. An example is:
DNA (3' end) ...CACCTACGTTCAGGAGGTCAGGACTGGTAC... (5' end)
(5' end) ...GTGGATGCAAGTCCTCCAGTCCTGACCATG... (3' end)
DNA is transcribed into mRNA starting at the 3' end of the DNA strand (the mRNDA begins at the 5' end). Each nucleotide of the DNA strand attracts its complementary RNA base through a pairing rule. mRNA is composed of the same bases except adenine (A) is swapped out for uracil and all the bases flip:
DNA -> mRNA
A -> U
T -> A
G -> C
C -> G
Doing transcription on the strand up top gives:
DNA (3' end) ...CACCTACGTTCAGGAGGTCAGGACTGGTAC... (5' end)
mRNA (5' end) ...GUGGAUGCAAGUCCUCCAGUCCUGACCAUG... (3' end)
Rapid Testing for COVID-19 & DMD
Let's look at how PCR testing works. When a gene is expressed, many thousands of copies of RNA are made per one copy of DNA. A test working with RNA would be better suited as a result, as we have more test sites to work with.
A retrovirus like HIV contains RNA rather than DNA. But these viruses carry reverse transcriptase which can turn single stranded RNA back into double stranded DNA.
PCR essentially uses Week 6 - Biotechnology#Genetic Testing - PCR and Gel Electrophoresis as we've talked about before.
For Week 6 - Biotechnology#Diagnosing DMD, we can do something similar. Using Lecture 6 - Finishing Genetics & DNA#RNA Processing, recall that exons are parts of the RNA that stay after the RNA is copied from the DNA. introns are the parts that get removed.
There is a new exciting approach to treat DMD using exon skipping, where we try to remove the DMD mutation by just skipping the exon that encodes the protein deficiency.
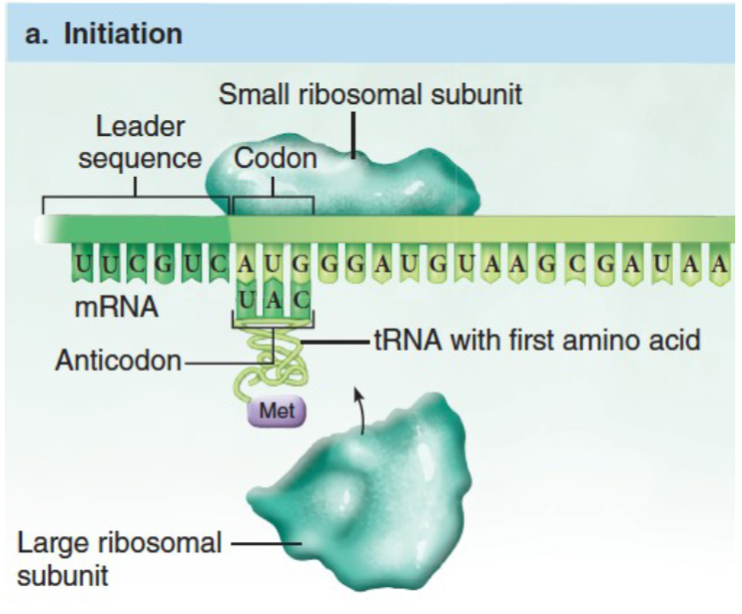
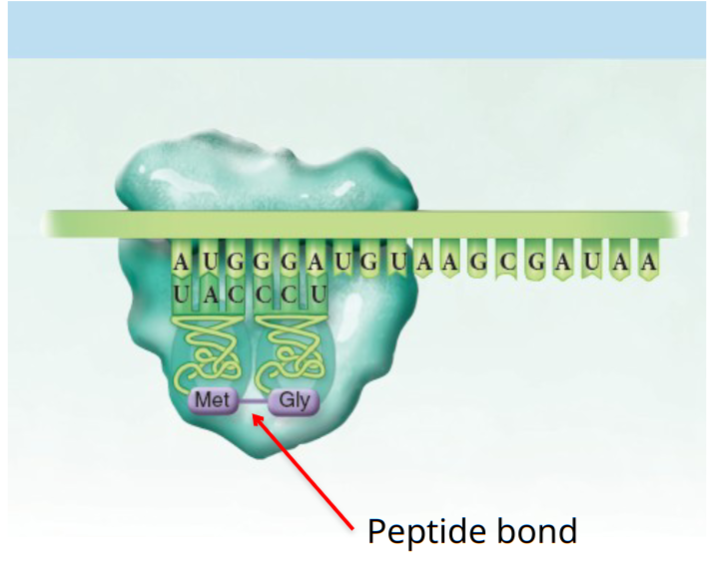
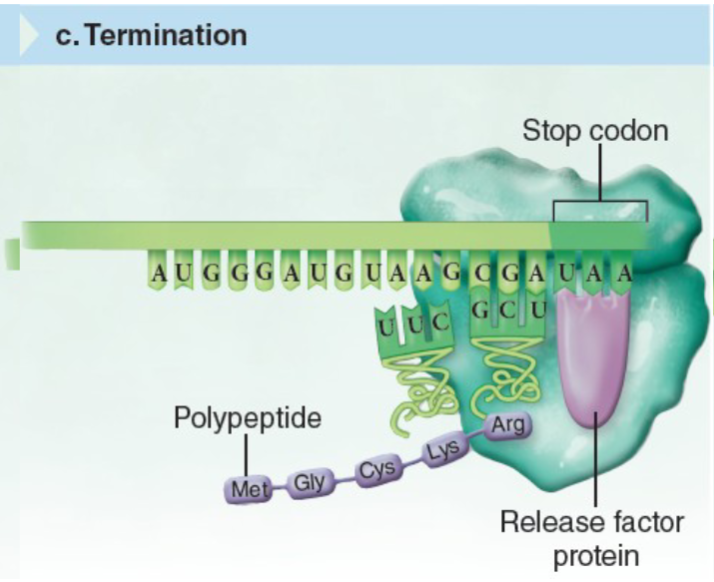
Translation
mRNA is translated into a sequence of amino acids by ribosomes. The ribosome attaches to the 5' end of the mRNA and slides towards the 3' side. Each codon in the mRNA triggers the ribosome to build a polypeptide chain until a STOP codon is reached.
See:
For reference:
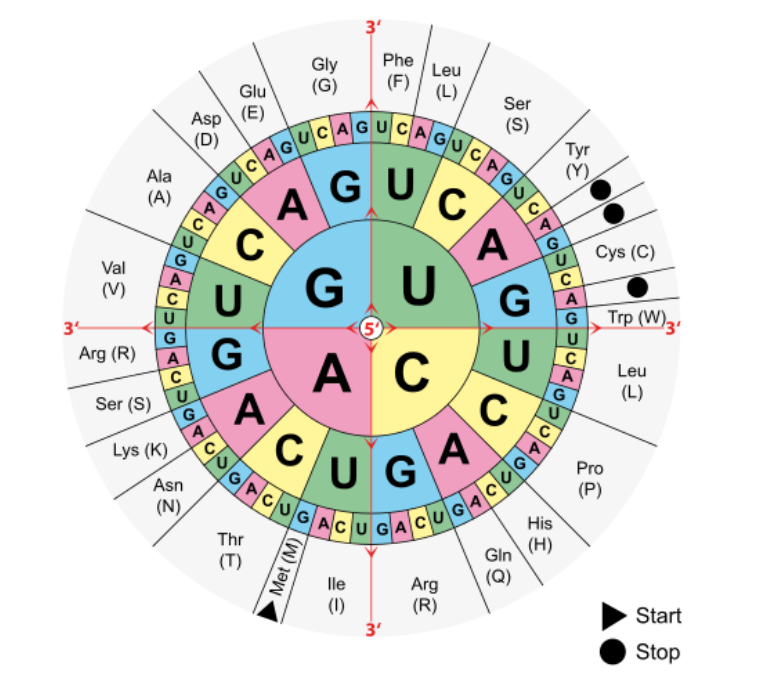
The steps are:
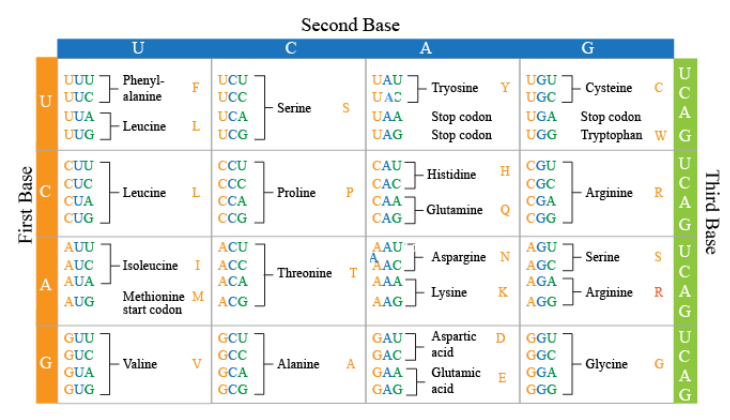
- Find the start codon
AUGstarting from5' - Read the sequence 3 letters at a time (each codon), building the polypeptide as you go. Include
Mfrom theAUGstart codon. - Keep reading until you hit one of the STOP codons.
As an example:
start codon v
mRNA (5' end) GUGGAUGCAAGUCCUCCAGUCCUGACCAUG (3' end)
mRNA triplets AUG CAA GUC CUC CAG UCC UGA
(start) (stop)
Amino acids: M Q V L Q S
Notice the duplication of some usages of some amino acids in the chart. These repetitions protect from point mutations which are discussed in more detail here.
Given the mRNA strand CUAUGGCCAUACGGUAGC let's produce:
CUA UGG CCA UAC GGU AGC
(ign) (ign) (ign) (ign) (ign) (ign)
(leu) (trp) (pro) (tyr) (gly) (ser)
... AUG GCC AUA CGG UAG ...
... (go) ... (stop)
... M A I R
Properties of Amino Acids
Each amino acid has a combination of chemical properties that makes it desirable in each protein we want to make. Substitutions of proteins that have similar properties will be less drastic to the function/form of the protein.
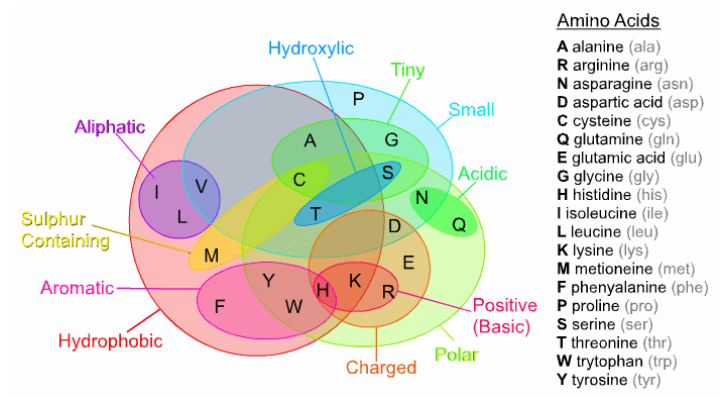
How much do these properties matter?
Let's consider the complex protein hemoglobin. It's a macromolecule that transports oxygen from the lungs to the tissues and carries
- Two alpha chains
- Two beta chains
Sickle cell anemia is a disease caused by a single nucleotide polymorphism, or change in a single base in the DNA. The 6th amino acid in the polypeptide chain is normally glu or E while people with the disease have valine val or V in the same position.
The subsitution turns the hydrophilic glutamic into hydrophobic valine, giving the tendency for the protein to collapse onto itself.

This data is in chromosome 11, so if both parent are unaffected carriers, there's a 1 in 4 chance that their offspring has the mutation of both chromosomes and thus has the condition.
Evolution and Bioinformatics
Bioinformatics naturally uses the fields of CSC and information technology. By studying many samples of genetic material, we can estimate the probability that mutations to the genetic code will cause one amino acid in a protein to be changed in a different amino acid. This is quantified by a substitution matrix. In the matrix below, each entry is a log-odds score, the log of a certain amino acid substitution taking place in a fixed number of generations. A positive number represents a common substitution. A negative number is less common. The numbers are denoted the PAM Score.
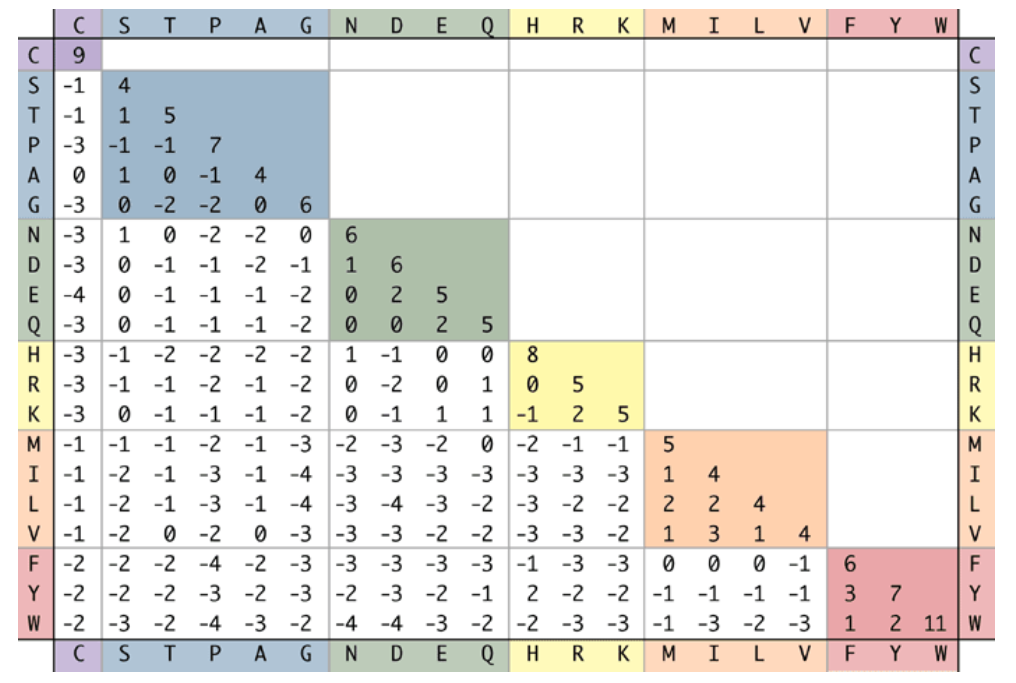
To compare a sequence of amino acids in two related proteins:
- 1: PEYDLLV
- 2: PERDILV
To get from
is a score of 7 is 5 - ...
Repeat and we get:
PAM score: +7 +5 -2 +6 +2 +4 +4 = 26
The lower the PAM score, the less likely the sequence progression from (1)
- The progression from (1)
(3) is less likely than - More of sequence (2) get's synthesized from (1) than (3).
We still try to align the sequences based on commonalities; however, any insertion or deletion itself gets an additional score of -5 added. You then have to consider all the places where the insertion/deletion occurs, and try to find the highest PAM score and assign that.
To use PAM scores to assess different progressions of evolution:
- Isolate and decode a particular gene in two different species
- Align the base pairs for the two genes
- Determine the sequence of amino acids coded by the genes
- Calculate a quantitative measure of the differences between the proteins produces by the two genes (ie: use the PAM score)
- Asses this difference in evolutionary terms
The short of it is that:
- Use the highest PAM score to indicate likelihood that evolution happened on that path (two species are more or less related via the PAM score)
- For instance, if
A/B = +12andA/C = +25andB/C = +8then- The probability of
is a score of - On contrast, the score for
directly is just , so it's more likely species came directly from rather than from who evolved species into .
- The probability of
Converting from WHEEL to WHIRL has a PAM score of:
W,W = +11
H,H = +8
E,I = -3
E,R = 0
L,L = 4
Giving a total PAM score of +11 + 8 - 3 + 0 + 4 = +20.